Dharshna Priya
Advisors: Dra. Aline Ramos Maia Lobba e Dr. Milton Yutaka Nishiyama-Jr.
Collaborator: Dr. Ana Claudia Carreira
Graduated in microbiology from the Faculty Of Arts and Sciences at the Mother Teresa University (Theni, India). Master’s degree in bioinformatics from Bharathidasan University (Tiruchirappalli, India) in conformational analysis of molecules. Doctoral student at the Postgraduate Program in Domestic and Wild Animals Anatomy at the Faculty of Veterinary Medicine and Animal Science at the University of São Paulo (USP). Currently working in bioinformatics applied to cellular and molecular biology and developing a project to characterize the role of CD90/Thy1 in triple-negative breast cancer (TNBC) through omic tools (RNAseq and Phospho-Proteomics) and bioinformatics
Analysis of signaling pathways in triple-negative breast cell lines overexpressing the CD90/Thy-1 gene (MCF10A/CD90+) or silenced (Hs578T/shCD90) through bioinformatics and proteomics approaches.
Breast cancer is the most prevalent cancer among women. Among the breast cancer subtypes, the triple-negative (TNBC) basal type is the most aggressive and with the worst prognosis within the ductal carcinoma subtype. Due to the lack of adequate molecular targets, the diagnosis and treatment of patients with the TNBC phenotype have been a major challenge. To understand the biology of TNBC, the molecular target CD90/Thy-1 was identified in our laboratory as being highly expressed in the basal-type TNBC cell line Hs578T, which is highly malignant and, on the other hand, has low expression in the MCF10A non-malignant
line (Lobba et al., 2012 Cytometry).
Based on the functional genomic approach, the CD90 expression was increased in the nonmalignant epithelial cell line (MCF10A) and silenced in the highly malignant cell line
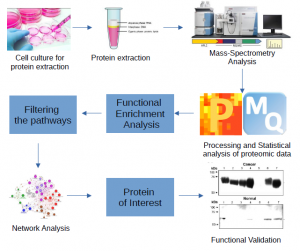
(Hs578T), allowing to demonstrate that the CD90 gene is involved in several cellular processes that lead to malignant transformation. Therefore, our study demonstrated that the CD90 gene can be considered a promising tumor marker of TNBC. In this context, this project aims to analyze the CD90 signaling pathways, based on literature data and databases, and confirm, through RNAseq and proteomics, the altered signaling pathways of genes and proteins. All data is analyzed using downstream bioinformatics analysis (MaxQuant and Perseus) to find differentially expressed proteins. Based on the data obtained, we suggest the regulation of processes such as cell migration, metastasis, and chemoresistance associated with the CD90 gene in TNBC.
This project may contribute to the understanding of altered pathways correlated with CD90 in order to seek new targets for the diagnosis and treatment of this type of triple-negative breast tumor.